Randomly prioritize conservation projects under expected phylogenetic diversity
Source:R/ppp_random_phylo_solution.R
ppp_random_phylo_solution.RdGenerate random solutions for the 'Project Prioritization Protocol'
problem (Joseph, Maloney & Possingham 2009) and evaluate them using
'expected phylogenetic diversity' (Faith 2008).
Although conservation projects should, ideally, not be funded based on random
allocations, it can be useful to compare the effectiveness of solutions to
random decisions in order to evaluate their effectiveness.
When informing conservation actions, it is strongly recommended to
use the ppp_exact_phylo_solution method because it can identify
optimal funding schemes with a guarantee.
ppp_random_phylo_solution(x, y, tree, budget, project_column_name, success_column_name, action_column_name, cost_column_name, locked_in_column_name = NULL, locked_out_column_name = NULL, number_solutions = 1L)
Arguments
| x |
|
|---|---|
| y |
|
| tree |
|
| budget |
|
| project_column_name |
|
| success_column_name |
|
| action_column_name |
|
| cost_column_name |
|
| locked_in_column_name |
|
| locked_out_column_name |
|
| number_solutions |
|
Details
Each random solution is generated using the following algorithm. Firstly, all actions are initially selected for funding (excepting actions which are locked out). Secondly, an action is randomly selected and defunded. Thirdly, the second step is repeated until the total cost of the remaining actions that are prioritized for funding is within the budget. Note that actions that have zero cost are never deselected for funding, and are always included in the solutions. Additionally, actions that are locked in are never deselected for funding.
References
Faith DP (2008) Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conservation Biology, 22: 1461--1470.
Joseph LN, Maloney RF & Possingham HP (2009) Optimal allocation of resources among threatened species: A project prioritization protocol. Conservation Biology, 23, 328--338.
See also
For other methods for generating solutions for the 'Project
Prioritization Protocol' problem using phylogenetic data, see
ppp_heuristic_phylo_solution
ppp_exact_phylo_solution, and
ppp_manual_phylo_solution.
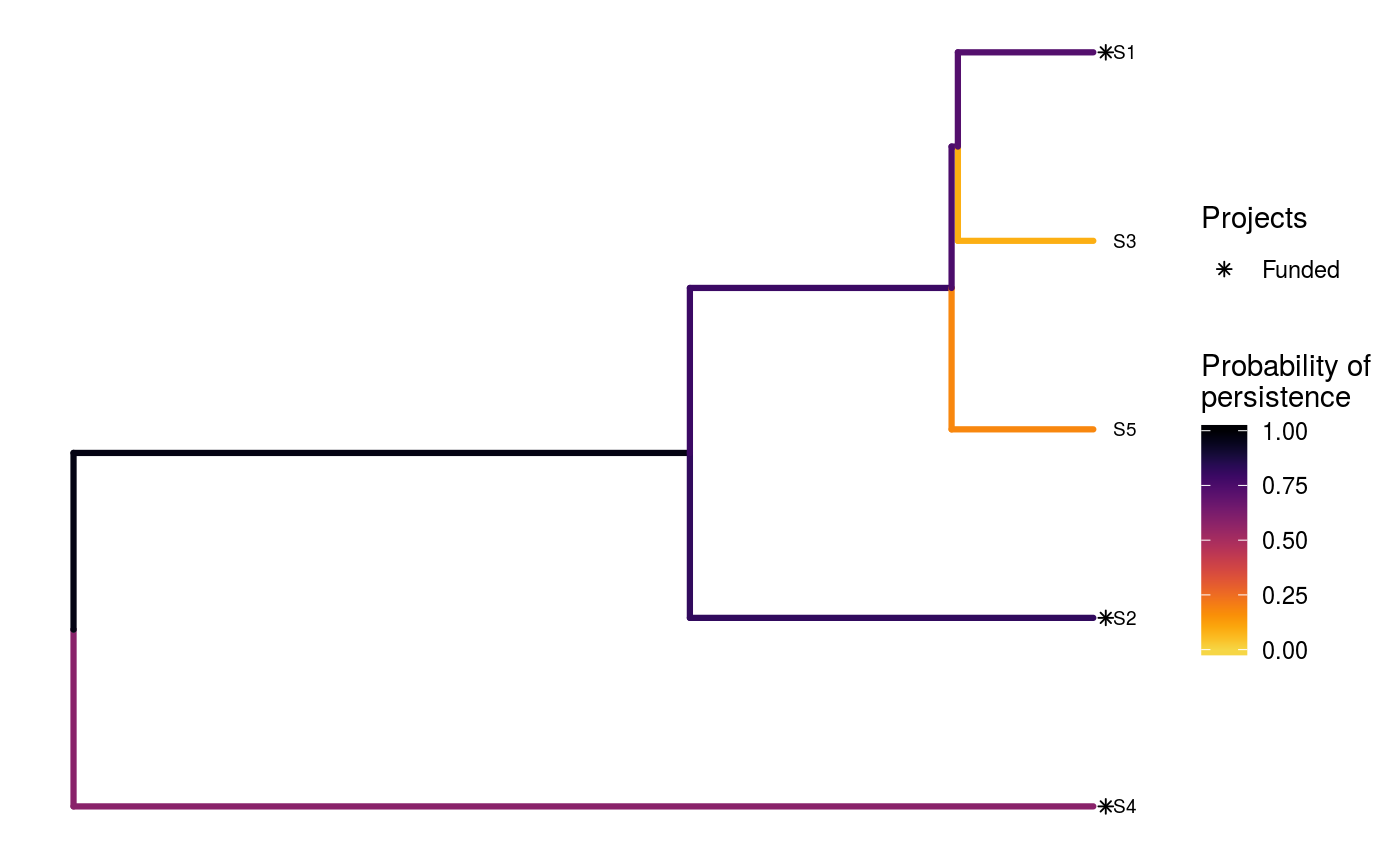
To visualize the effectiveness of a particular solution, see
ppp_plot_phylo_solution.
Examples
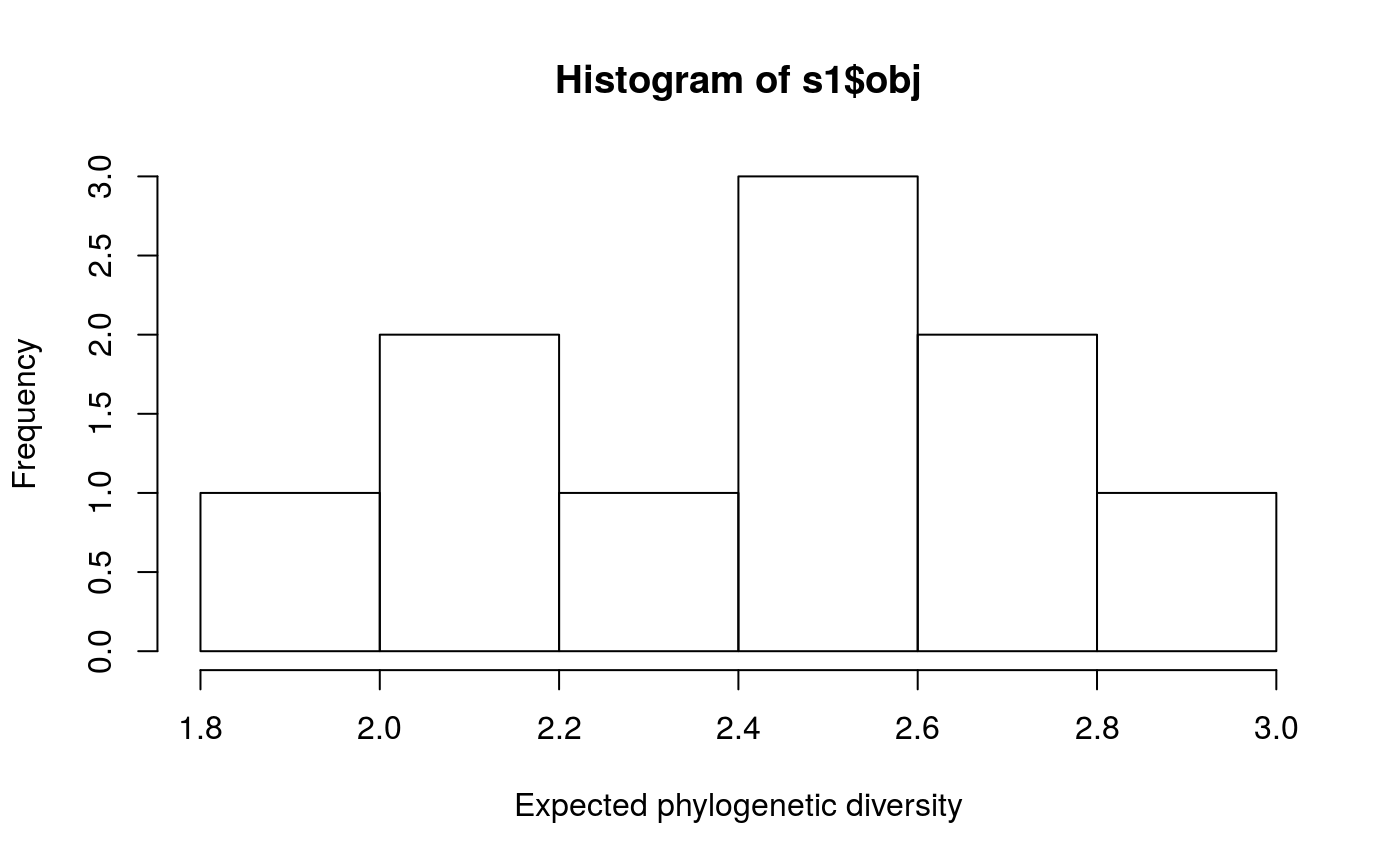
# set seed for reproducibility set.seed(500) # load built-in data data(sim_project_data, sim_action_data, sim_tree) # print simulated project data print(sim_project_data)#> # A tibble: 6 x 13 #> name success S1 S2 S3 S4 S5 S1_action S2_action S3_action #> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl> <lgl> #> 1 S1_p~ 0.919 0.791 0 0 0 0 TRUE FALSE FALSE #> 2 S2_p~ 0.923 0 0.888 0 0 0 FALSE TRUE FALSE #> 3 S3_p~ 0.829 0 0 0.502 0 0 FALSE FALSE TRUE #> 4 S4_p~ 0.848 0 0 0 0.690 0 FALSE FALSE FALSE #> 5 S5_p~ 0.814 0 0 0 0 0.617 FALSE FALSE FALSE #> 6 base~ 1 0.298 0.250 0.0865 0.249 0.182 FALSE FALSE FALSE #> # ... with 3 more variables: S4_action <lgl>, S5_action <lgl>, #> # baseline_action <lgl>#> # A tibble: 6 x 4 #> name cost locked_in locked_out #> <chr> <dbl> <lgl> <lgl> #> 1 S1_action 94.4 FALSE FALSE #> 2 S2_action 101. FALSE FALSE #> 3 S3_action 103. TRUE FALSE #> 4 S4_action 99.2 FALSE FALSE #> 5 S5_action 99.9 FALSE TRUE #> 6 baseline_action 0 FALSE FALSE#> #> Phylogenetic tree with 5 tips and 4 internal nodes. #> #> Tip labels: #> [1] "S3" "S1" "S5" "S2" "S4" #> #> Rooted; includes branch lengths.# generate 10 random solutions that meet a budget of 300 s1 <- ppp_random_phylo_solution(sim_project_data, sim_action_data, sim_tree, 300, "name", "success", "name", "cost", number_solutions = 10) # print solutions print(s1)#> # A tibble: 10 x 12 #> solution method obj budget cost optimal S1_action S2_action S3_action #> <int> <chr> <dbl> <dbl> <dbl> <lgl> <lgl> <lgl> <lgl> #> 1 1 random 2.92 300 295. NA TRUE TRUE FALSE #> 2 2 random 2.60 300 297. NA TRUE FALSE TRUE #> 3 3 random 2.64 300 200. NA FALSE TRUE FALSE #> 4 4 random 2.50 300 299. NA TRUE TRUE TRUE #> 5 5 random 2.20 300 298. NA TRUE FALSE TRUE #> 6 6 random 2.50 300 299. NA TRUE TRUE TRUE #> 7 7 random 1.96 300 203. NA FALSE FALSE TRUE #> 8 8 random 2.50 300 295. NA TRUE TRUE FALSE #> 9 9 random 2.20 300 298. NA TRUE FALSE TRUE #> 10 10 random 2.28 300 202. NA FALSE FALSE TRUE #> # ... with 3 more variables: S4_action <lgl>, S5_action <lgl>, #> # baseline_action <lgl># plot first solution ppp_plot_phylo_solution(sim_project_data, sim_action_data, sim_tree, s1, "name", "success", "name", "cost", n = 1)# view histogram of their expected phylogenetic diversity hist(s1$obj, xlab = "Expected phylogenetic diversity")