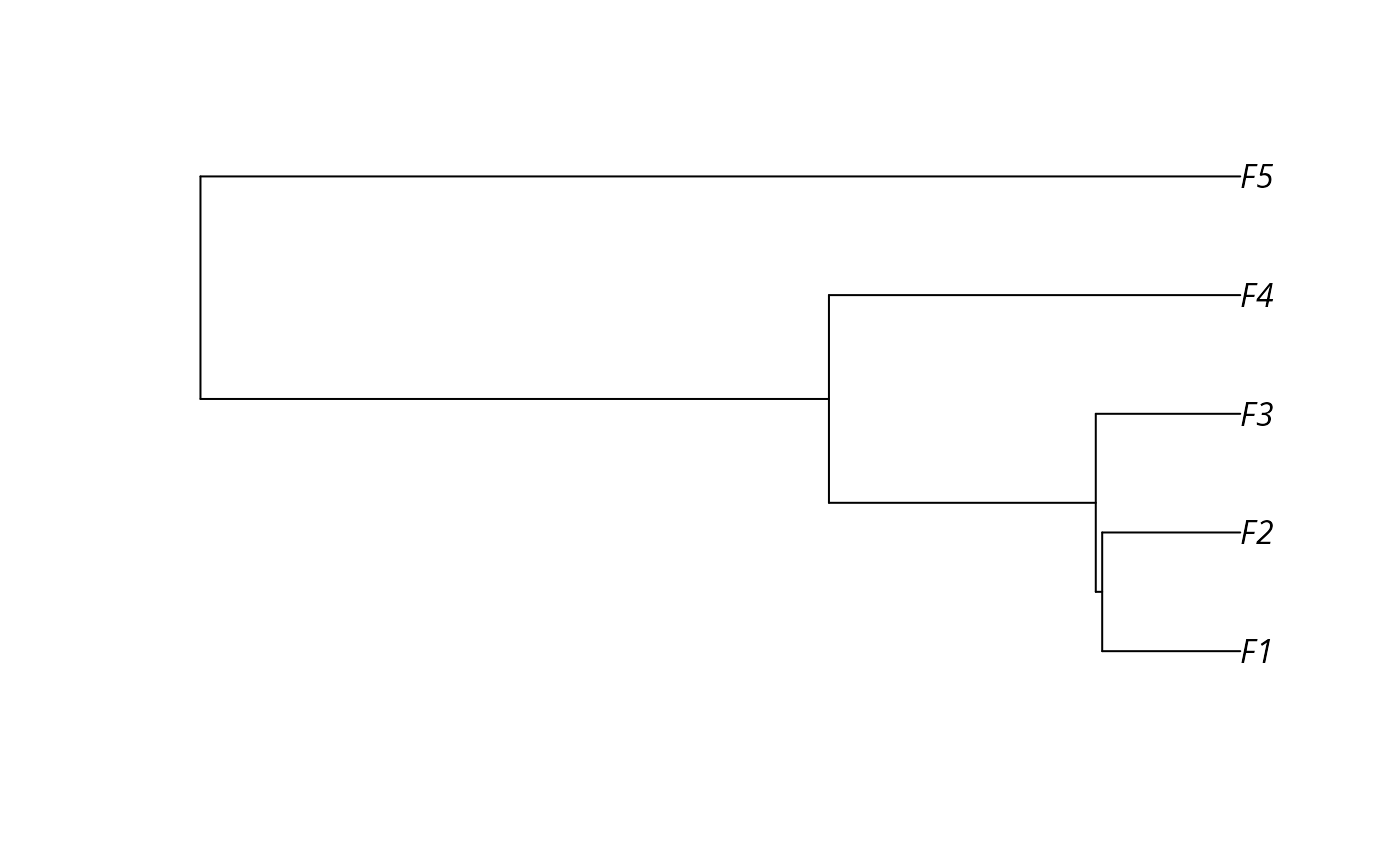
Simulated data for prioritizing conservation projects.
Format
- sim_projects
tibble::tibble()object.- sim_actions
tibble::tibble()object.- sim_features
tibble::tibble()object.- sim_tree
ape::phylo()object.
Details
The data set contains the following objects:
sim_projectsA
tibble::tibble()object containing data for six simulated conservation projects. Each row corresponds to a different project and each column contains information about the projects. This table contains the following columns:"name"charactername for each project."success"numericprobability of each project succeeding if it is funded."F1"..."F5"numericcolumns for each feature (i.e."F1","F2","F3","F4","F5", indicating the enhanced probability that each feature will survive if it funded. Missing values (NA) indicate that a feature does not benefit from a project being funded."F1_action"..."F5_action"logicalcolumns for each action, ranging from"F1_action"to"F5_action"indicating if an action is associated with a project (TRUE) or not (FALSE)."baseline_action"logicalcolumn indicating if a project is associated with the baseline action (TRUE) or not (FALSE). This action is only associated with the baseline project.
sim_actionsA
tibble::tibble()object containing data for six simulated actions. Each row corresponds to a different action and each column contains information about the actions. This table contains the following columns:"name"charactername for each action."cost"numericcost for each action."locked_in"logicalindicating if certain actions should be locked into the solution."locked_out"logicalindicating if certain actions should be locked out of the solution.
sim_featuresA
tibble::tibble()object containing data for five simulated features. Each row corresponds to a different feature and each column contains information about the features. This table contains the following columns:"name"charactername for each feature."weight"numericweight for each feature.
- tree
ape::phylo()phylogenetic tree for the features.
Examples
# load data
data(sim_projects, sim_actions, sim_features, sim_tree)
# print project data
print(sim_projects)
#> # A tibble: 6 × 13
#> name success F1 F2 F3 F4 F5 F1_action F2_action
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl>
#> 1 F1_project 0.919 0.791 NA NA NA NA TRUE FALSE
#> 2 F2_project 0.923 NA 0.888 NA NA NA FALSE TRUE
#> 3 F3_project 0.829 NA NA 0.502 NA NA FALSE FALSE
#> 4 F4_project 0.848 NA NA NA 0.690 NA FALSE FALSE
#> 5 F5_project 0.814 NA NA NA NA 0.617 FALSE FALSE
#> 6 baseline_proj… 1 0.298 0.250 0.0865 0.249 0.182 FALSE FALSE
#> # ℹ 4 more variables: F3_action <lgl>, F4_action <lgl>, F5_action <lgl>,
#> # baseline_action <lgl>
# print action data
print(sim_actions)
#> # A tibble: 6 × 4
#> name cost locked_in locked_out
#> <chr> <dbl> <lgl> <lgl>
#> 1 F1_action 94.4 FALSE FALSE
#> 2 F2_action 101. FALSE FALSE
#> 3 F3_action 103. TRUE FALSE
#> 4 F4_action 99.2 FALSE FALSE
#> 5 F5_action 99.9 FALSE TRUE
#> 6 baseline_action 0 FALSE FALSE
# print feature data
print(sim_features)
#> # A tibble: 5 × 2
#> name weight
#> <chr> <dbl>
#> 1 F1 0.211
#> 2 F2 0.211
#> 3 F3 0.221
#> 4 F4 0.630
#> 5 F5 1.59
# plot phylogenetic tree
plot(sim_tree)